High-resolution HLA typing
1.Background
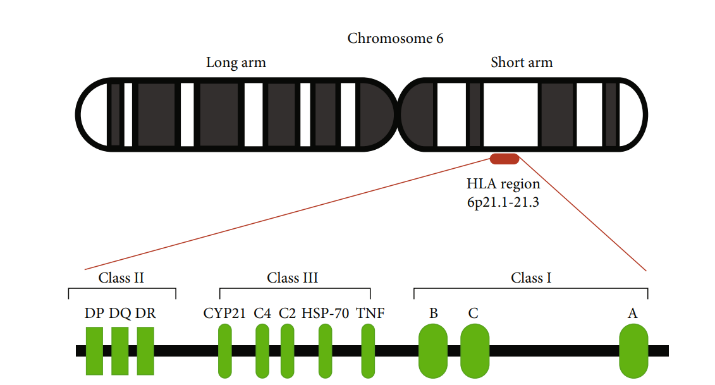
HLAs (Human Leukocyte Antigens) are products expressed by the human Major Histocompatibility Complex (MHC) genes. These molecules are involved in antigen presentation, regulating intercellular protein recognition and inducing immune responses. HLAs are divided into three main categories: classical HLA class I (HLA-A, B, C), classical HLA class II (HLA-DP, DQ, DR), and non-classical HLA.
HLA molecules are the most diverse group of genes in humans. They exhibit extensive polymorphism in human individuals to ensure the broad adaptability and resistance of the human population to various pathogens. The inheritance and expression of HLA genes are related to all aspects of our health. Certain HLA alleles are involved in or even cause the occurrence and development of almost all autoimmune diseases, and many allergic diseases, infectious diseases, and metabolic diseases. For instance, psoriasis is associated with HLA-C*06:02, type I diabetes with HLA-DQ2 and HLA-DQ8, ankylosing spondylitis with HLA-B27, rheumatoid arthritis with HLA-DR4, and systemic lupus erythematosus with HLA-DR3.
Researchers in the field of immunotherapy may pay more attention to the role of HLA genes in treatment. A patient's HLA typing not only affects the success rate of their immunotherapy but also determines whether they can receive some highly personalized cell or antibody treatments, such as TCR-T therapy and TCRm-ADC. Therefore, in the scientific research of T-cell immunity and related drug development, the HLA genotype of the donor is also essential information to know.
2.Nomenclature
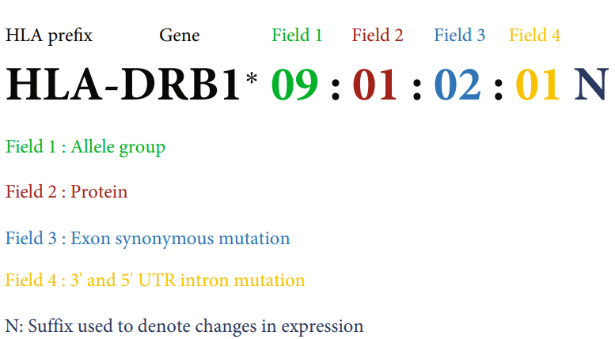
The nomenclature of HLA genes follows the format HLA-gene name*serological group: identifier for specific protein: identifier for exon nucleotide sequence: identifier for intron and UTR mutations. For example, HLA-A*01:01:01:01 and HLA-A*01:01:02 express identical proteins, but there are synonymous mutations in the exons encoding the proteins. Generally, in the fields of bone marrow transplantation or gene therapy, the precision requirement for HLA typing is 2 or 3 fields. The above two genes are referred to as HLA-A*01:01:01 and HLA-A*01:01:02, both belonging to HLA-A*01:01. Some genotypes contain letters, such as N, S, etc., which represent different meanings. For example, N indicates a coding frame shift, and the gene cannot express a complete HLA protein. S indicates that the HLA will be expressed as a secreted protein rather than a membrane protein.
3.Methods
Currently, there are four main methods for detecting HLA typing.
-
SBT typing method based on first-generation Sanger sequencing:
The Sanger sequencing method is considered the gold standard by current researchers. The method involves specific amplification of HLA genes and analysis of results using first-generation sequencing. The advantage of this method is high accuracy, which stems from Sanger sequencing's inherent 99.99% base calling accuracy rate. Its limitations include: data analysis requiring special programs, and multi-locus typing work requires separate workflows, resulting in relatively low throughput. Additionally, Exons 5, 6, 7 for type I and Exons 4, 5 for type II HLA are typically not typed. In rare cases, frameshift or other mutations may lead to nullified or abnormal expression of the molecules compared to be expected from the genotyping results.
-
NGS typing method based on second-generation sequencing:
The NGS typing method is also based on target gene enrichment, sequencing and data analysis. Since the single-base accuracy of NGS is slightly inferior to Sanger sequencing, and earlier typing algorithms lacked precision, researchers have not adopted the NGS method as the gold standard. However, with improvements in NGS technology bringing increased single-base accuracy, improved timeliness, and reduced costs, NGS is becoming a method that balances timeliness, accuracy, and cost in the future.
-
Typing method based on third-generation sequencing:
Third-generation sequencing-based typing methods, while ensuring accuracy, can resolve some technical difficulties that occur in first-generation sequencing, such as phase ambiguity at certain exon positions. However, this technology is currently limited by the cost issues of third-generation sequencing technology itself, resulting in relatively low adoption rates.
-
rSSO typing method based on Luminex platform:
Taking advantage of the Luminex platform, it can quickly render intermediate to high-resolution typing results, with the slight disadvantage of being unable to identify rare alleles and not covering the full gene length.
4.Advantages
Since 2020, we have been developing HLA typing methods. We have obtained invention patents based on Sanger sequencing methods. We have provided HLA typing services for many enterprises and research institutions, maintaining very high client satisfaction. Tiger Cellular Remedy has also developed efficient NGS based typing technology, accurately presenting results with 3-fields high resolution. TigerHLA, our proprietary algorithm can greatly improve accuracy compared to traditional open-source algorithms (see examples). By simultaneously having Sanger sequencing based and NGS based typing methods for cross-validation, we can ensure a very high accuracy rate. Additionally, our NGS typing method is based on probe enrichment, effectively avoiding the PCR artifacts and allelic dropout errors associated with long range PCR.
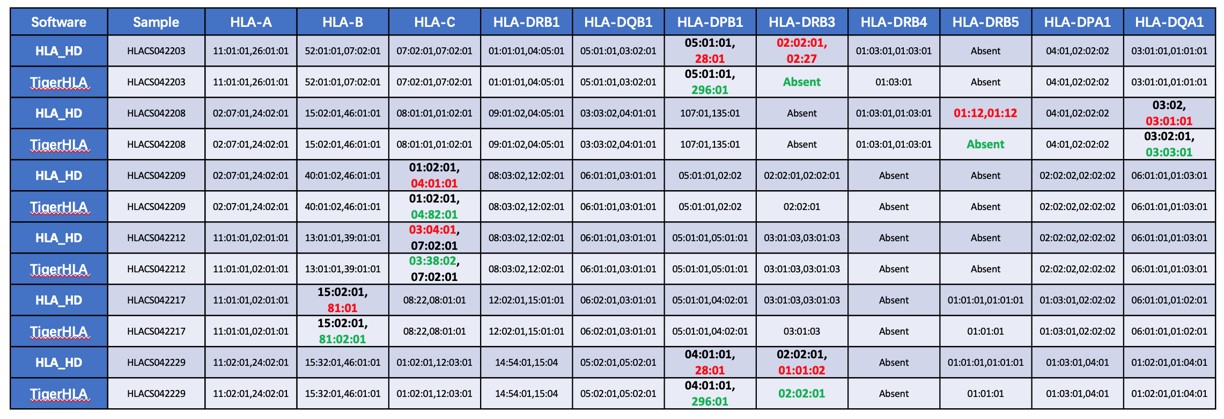 All samples above were confirmed by Sanger sequencing.Processing this dataset (35 samples, 385 locus in total), the TigerTCR HLA typing software showed 100% concordance, whereas open source tools (e.g., HLA-HD) presented erroneous results (97-98% overall accuracy on loci HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DPB1, HLA-DQA1, HLA-DPA1 and lower than 80% accuracy on loci HLA-DRB3/4/5)
All samples above were confirmed by Sanger sequencing.Processing this dataset (35 samples, 385 locus in total), the TigerTCR HLA typing software showed 100% concordance, whereas open source tools (e.g., HLA-HD) presented erroneous results (97-98% overall accuracy on loci HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DPB1, HLA-DQA1, HLA-DPA1 and lower than 80% accuracy on loci HLA-DRB3/4/5)
5.Pricing
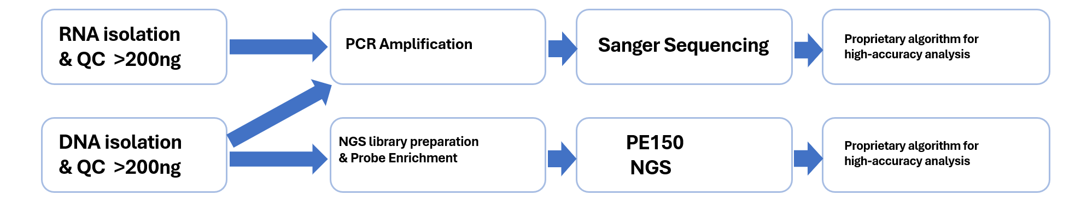
1.Method Process
2.Detection Range
Currently, we provide HLA typing services based on NGS and SBT methods. The NGS method covers 11 loci, HLA - A, B, C, DRB1, DQB1, DPB1, DQA1, DPA1, DRB3,4,5. The SBT method covers 6 locis, including HLA - A, B, C, DRB1, DQB1, DPB1. For international samples:
- SBT Allele Typing – 99 USD/sites/sample
- NGS 6-locus typing (HLA-A, B, C, DRB1, DQB1, DPB1) – 139 USD/sample
- NGS 11-locus typing (HLA-A, B, C, DRB1, DQB1, DPB1, DQA1, DPA1, DRB3,4,5) – 159 USD/sample
Additional one-time shipping cost and custom clearance cost will apply, regardless of the sample count.
